Online multi-omics data integration workshop series
January 2021
In the first months of 2021, an online X-omics workshop series took place. Different aspects of challenges and solutions related to multi-omics data integration were being addressed in four workshops.
Workshop program
 Workshop #1: Multi-omics helpdesk, data-centric analysis platform and NGS diagnostics case study – January 20, 2021, 15:00 – 17:00 CET
Workshop #1: Multi-omics helpdesk, data-centric analysis platform and NGS diagnostics case study – January 20, 2021, 15:00 – 17:00 CET
- Daniella Kasteel (X-omics): "Access to the X-omics helpdesk".
The X-omics helpdesk can give you advice on a X-omics approach, on specific omics related sample or data analysis challenges and help you optimize your study design. In her talk, Daniella will highlight the application process and the support the X-omics helpdesk is offering researchers.Access to the X-omics helpdesk
- Leon Mei (LUMC): "Data-centric multi-omics analysis platform used in the BBMRI-NL project".
For BBMRI-NL researchers, all data shared by participating biobanks including genome-wide SNP data, limited phenotypes, RNA-seq and DNA methylation on all individuals can be analyzed on centralized computational facilities, namely the BIOS VM hosted at the SurfSARA High Performance Computing cloud. To provide a more flexible and scalable analysis platform, we have started using the new Research Science Cloud at SURFsara. In this talk, Leon will highlight the architecture, on-boarding procedure and lessons learned using this new infrastructure for future centralized multiomics data analysis within the BBMRI-NL community.Data centric multi-omics analysis platform used in the BBMRI-NL project
- Joeri van der Velde & Lennart Johansson (UMCG): "Cutting-edge NGS diagnostics: from interpretation to FAIRification & challenges of large-scale Rare Disease cohort analysis".
In this joint talk, Joeri will discuss the latest developments of computational tools and pipelines for genome diagnostics, as well as FAIRification of NGS data. Lennart will highlight the challenges and potential of analyzing NGS data in a cohort of 19,000 Rare Disease patients.
 Workshop #2: Multi-omics integration tools and platforms – February 10, 2021, 15:00 – 17:00 CET
Workshop #2: Multi-omics integration tools and platforms – February 10, 2021, 15:00 – 17:00 CET
We will have a look at the different tools and platforms mixOmics, cBioPortal and R2.
Watch recorded workshop 2
- Remond Fijneman (NKI): "Access, query and view your translational research study in cBioPortal: the MEDOCC use case"
Translational research studies face significant challenges with respect to data management and data analysis. Longitudinal clinical, imaging, biobanking and molecular data need to be collected and integrated to access, query and view translational research data. cBioPortal is an open-access, open-source resource for interactive exploration of multidimensional (cancer) genomics data sets. The cBioPortal tool is adopted and supported by Health-RI to facilitate multicenter translational research studies that are initiated in the Netherlands. One of these studies is PLCRC-MEDOCC, referring to Molecular Early Detection Of Colorectal Cancer (MEDOCC) as a substudy of the national prospective colorectal cancer cohort study (PLCRC). The aim of the PLCRC-MEDOCC study is to investigate cell-free circulating tumor DNA (ctDNA) as a biomarker for minimal residual disease in stage II and stage III colon cancer. The MEDOCC use case will be presented as an example of an international multicenter translational research study for which longitudinal clinical, biobanking and molecular data are currently being collected and integrated. - Jan Koster (Amsterdam UMC): "The R2 Genomics Analysis and Visualization Platform: Illustration of a multi omics journey into the unraveling of cancer biology"
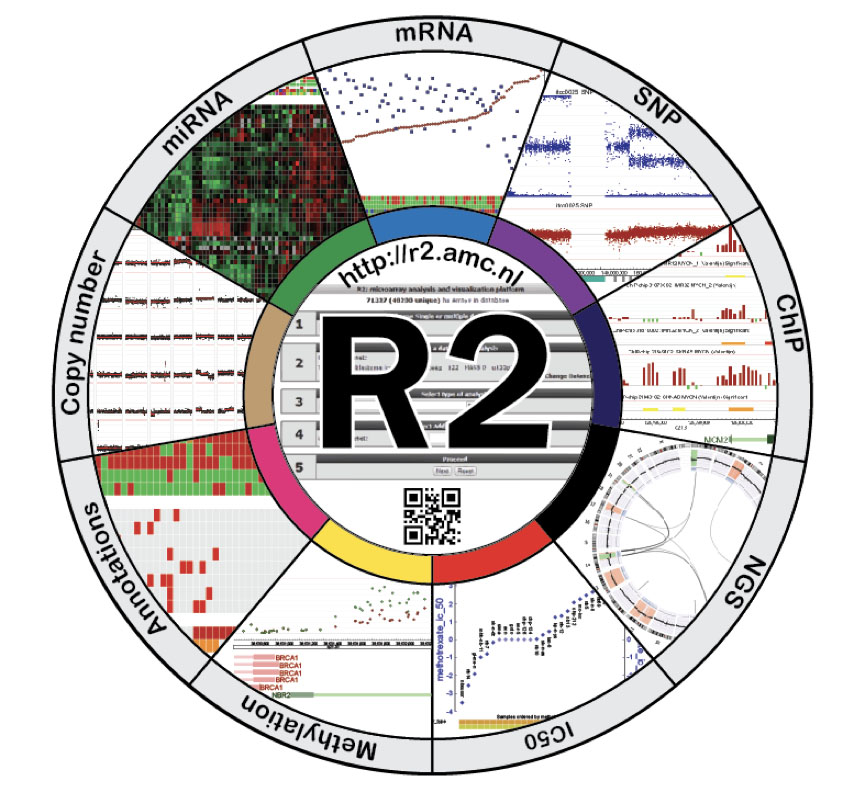 In this short talk, Jan Koster (who conceived and developed the R2 platform) will be taking you on a little journey to unravel fundamental aspects in cancer biology, using the pediatric cancer, neuroblastoma. The R2 Genomics Analysis and Visualization Platform (r2.amc.nl) has been originally designed and further developed, closely linked with this journey, while it was also being discovered by new users. We will touch on some historic findings, see the introduction of multi omics data sets in the daily practice and illustrate this on some analyses and visualizations that have been co-developed in R2 to assist and accompany researches on their quest to find clues to stop cancer.
In this short talk, Jan Koster (who conceived and developed the R2 platform) will be taking you on a little journey to unravel fundamental aspects in cancer biology, using the pediatric cancer, neuroblastoma. The R2 Genomics Analysis and Visualization Platform (r2.amc.nl) has been originally designed and further developed, closely linked with this journey, while it was also being discovered by new users. We will touch on some historic findings, see the introduction of multi omics data sets in the daily practice and illustrate this on some analyses and visualizations that have been co-developed in R2 to assist and accompany researches on their quest to find clues to stop cancer. - Kim-Anh Le Cao (University of Melbourne): "Multivariate Data Integration with mixOmics"
Brief overview of the main concepts and methods implemented in mixOmics, which are based on Projection to Latent Structures algorithm. Some examples of applications in various omics data integration problems will be shown and the next methodological developments for the package will be briefly covered.
Access, query and view your translational research study in cBioPortal: the MEDOCC use case
Multivariate Data Integration with mixOmics
 Workshop #3: Predictive multi-omics biomarker signatures – February 24, 2021, 15:00 – 17:00 CET
Workshop #3: Predictive multi-omics biomarker signatures – February 24, 2021, 15:00 – 17:00 CET
The third workshop will highlight –omics clocks and –omics risk scores.
Watch recorded workshop 3
- Patrick Deelen (UMCU & UMCG): "Reconciling GWAS and Mendelian genetics through core-gene identification using Downstreamer"
I will show how we use genetics and RNA-seq to prioritize the important genes of common diseases and traits, such as auto-immine disorders and height. We observed that these prioritized genes are enriched for genes that cause rare diseases when damaged. This shows the shared biology of common and rare diseases. - Gennady Roshchupkin (Erasmus MC): "Artificial Intelligence and multi-omics data: Better, Faster, Stronger"
In this talk we will discuss the latest development in AI technologies (machine learning and deep learning) in application to various omics data. How such algorithms can help to analyze data and make new discoveries. What are the obstacles we need to overcome first and what are the new opportunities AI can bring to omics field? - Erik van den Akker (LUMC): "Metabolomics-based predictors as markers of health and disease"
The blood metabolome incorporates cues from the environment and the host’s genetic background, potentially offering a holistic view of an individual’s health status. Using vast resources of proton nuclear magnetic resonance metabolomics and phenotypic data we constructed predictors for mortality, biological age, and a wide array of cardio-metabolic variables routinely assessed in large epidemiological studies. For the mortality and biological age predictors we show associations with various clinical end points, suggesting that they pick up on a broad range of health aspects. We illustrate the value of the predictors for cardio-metabolic variables by using them to impute missing phenotypic data that can be employed for downstream association analyses.
Reconciling GWAS and Mendelian genetics through core-gene identification using Downstreamer
Artificial Intelligence and multi-omics data: Better, Faster, Stronger
Metabolomics-based predictors as markers of health and disease
 Workshop #4: Pitch your own research project – March 10, 2021, 15:00 – 17:00 CET
Workshop #4: Pitch your own research project – March 10, 2021, 15:00 – 17:00 CET
Finally, we invite you to pitch your own project and get advice from panelists and participants on experimental design and data integration strategies for a multi-omics approach.
Biosketches of our speakers
Daniella Kasteel
 Dr.Ir. Daniella Kasteel is the project manager of X-omics. She is an experienced project/research manager with a scientific background. Apart from the general project management tasks she is responsible for the marketing and communication of X-omics, organization of events and workshops, the X-omics helpdesk and building a X-omics community.
Dr.Ir. Daniella Kasteel is the project manager of X-omics. She is an experienced project/research manager with a scientific background. Apart from the general project management tasks she is responsible for the marketing and communication of X-omics, organization of events and workshops, the X-omics helpdesk and building a X-omics community.
Leon Mei
 Leon Mei is the head of Sequencing Analysis Support Core at LUMC. He was also involved in the BBMRI-GoNL project and chaired the data management group in the BBMRI-BIOS project.
Leon Mei is the head of Sequencing Analysis Support Core at LUMC. He was also involved in the BBMRI-GoNL project and chaired the data management group in the BBMRI-BIOS project.
Joeri van der Velde
 Kasper Joeri van der Velde graduated on pathway visualization of kinase activity at the Groningen Bioinformatics Centre in collaboration with the UMC Groningen dept. of Cell Biology, and subsequently worked as scientific programmer on web-based databases, methods and visualization tools for various multi-omics research projects involving plants, mice and nematodes. He then moved to the UMC Groningen dept. of Genetics to complete his PhD on translational software infrastructure for medical genetics in the group of Prof. Morris Swertz, where he is now a Postdoc. Using a unique combination of machine learning and data infrastructure research, he aims to discover new ways to revolutionarize the speed, yield and applications of medical genomics and bring those innovations into clinical practice where they can truly make a difference. In his work, he is a strong advocate of open source programming, open access science, and FAIR (findable, accessible, interoperable, reusable) data.
Kasper Joeri van der Velde graduated on pathway visualization of kinase activity at the Groningen Bioinformatics Centre in collaboration with the UMC Groningen dept. of Cell Biology, and subsequently worked as scientific programmer on web-based databases, methods and visualization tools for various multi-omics research projects involving plants, mice and nematodes. He then moved to the UMC Groningen dept. of Genetics to complete his PhD on translational software infrastructure for medical genetics in the group of Prof. Morris Swertz, where he is now a Postdoc. Using a unique combination of machine learning and data infrastructure research, he aims to discover new ways to revolutionarize the speed, yield and applications of medical genomics and bring those innovations into clinical practice where they can truly make a difference. In his work, he is a strong advocate of open source programming, open access science, and FAIR (findable, accessible, interoperable, reusable) data.
Lennart Johansson
 Lennart F. Johansson graduated as a BASc Life Sciences and Technology on the implementation of comparative genomic hybridization for the purpose of hematological malignancies at the University Medical Center Groningen (UMCG) department of genetics in 2002 and obtained a BA philosopy in 2015. In his thesis he formulated an answer to the question: ‘does new knowledge about existing results give a moral duty to recontact in clinical genetics?’. In 2019 he completed his PhD at the UMCG department of Genetics on the topic ‘Novel algorithms for genetic variation detection’ in which he describes the development of algorithms to be used on different NGS methods for copy number variation calling, translocation calling and NIPT. Next to a general discussion two extra reflection chapters on the epistemology and ethics of NGS finalized his thesis ‘Looking through the noise’. Currently he is a postdoc/project coordinator focusing on rare disease projects, such as EJP-RD, Solve-RD and CINECA, both on the side of infrastructure (HPC Computing, (meta-)databases) as data-analysis.
Lennart F. Johansson graduated as a BASc Life Sciences and Technology on the implementation of comparative genomic hybridization for the purpose of hematological malignancies at the University Medical Center Groningen (UMCG) department of genetics in 2002 and obtained a BA philosopy in 2015. In his thesis he formulated an answer to the question: ‘does new knowledge about existing results give a moral duty to recontact in clinical genetics?’. In 2019 he completed his PhD at the UMCG department of Genetics on the topic ‘Novel algorithms for genetic variation detection’ in which he describes the development of algorithms to be used on different NGS methods for copy number variation calling, translocation calling and NIPT. Next to a general discussion two extra reflection chapters on the epistemology and ethics of NGS finalized his thesis ‘Looking through the noise’. Currently he is a postdoc/project coordinator focusing on rare disease projects, such as EJP-RD, Solve-RD and CINECA, both on the side of infrastructure (HPC Computing, (meta-)databases) as data-analysis.
Remond Fijneman
 Remond Fijneman is Principal Investigator within the Translational Gastrointestinal Oncology group at the Department of Pathology, Netherlands Cancer Institute (Amsterdam, the Netherlands) with special interest in colorectal cancer translational research. Research activities are focused on identification and validation of diagnostic, prognostic, predictive and disease monitoring biomarkers for colorectal cancer by molecular profiling at DNA, RNA and protein level, for translation into clinical applications. At present, several cell-free circulating tumor DNA multicenter studies are carried out concerning detection of minimal residual disease in stage II and stage III colon cancer and monitoring treatment response in metastatic colorectal cancer. Significant effort is put in adopting and adapting the (inter)national translational research IT infrastructure to support and enable molecular research in the context of multicenter translational research projects.
Remond Fijneman is Principal Investigator within the Translational Gastrointestinal Oncology group at the Department of Pathology, Netherlands Cancer Institute (Amsterdam, the Netherlands) with special interest in colorectal cancer translational research. Research activities are focused on identification and validation of diagnostic, prognostic, predictive and disease monitoring biomarkers for colorectal cancer by molecular profiling at DNA, RNA and protein level, for translation into clinical applications. At present, several cell-free circulating tumor DNA multicenter studies are carried out concerning detection of minimal residual disease in stage II and stage III colon cancer and monitoring treatment response in metastatic colorectal cancer. Significant effort is put in adopting and adapting the (inter)national translational research IT infrastructure to support and enable molecular research in the context of multicenter translational research projects.
Jan Koster
 After having been educated as a molecular biologist (Utrecht University), and subsequently trained as a cell biologist at the Netherlands Cancer Institute (NKI) (supervised by Prof. A. Sonnenberg), Jan Koster switched career to the at that time developing field of bioinformatics in 2003. He joined the team of Prof. Versteeg, and in 2007 obtained a staff position within the department with his own team of bioinformaticians. His group currently consists of 6 bioinformaticians. The focus of Koster’s group is on applied bioinformatics, where they try to bridge the gap between the molecular biologists and the complex data that they are generating using high throughput technologies. Aside from helping the biologists with the processing, analysis and interpretation of HT data, they have heavily invested in an idea conceived in 2005. ‘Wouldn’t it be great if molecular biologists could analyze and explore their own data’. With this aim in mind and their biology background and mindset they have developed the web-based R2 genomic analysis and visualization platform (http://r2.amc.nl). R2 is composed of a dedicated database containing the HT data, coupled to a web-based suite of analysis and visualization tools that can be operated by users without bioinformatics training. R2 handles many types of genomics data from mRNA gene expression to whole genome sequencing data. The success of this concept can be illustrated by a user base of 140,000+ users (IP-addresses) from around the world, ~2,000,000 page views a year and over 1,500 citations in PubMed listed publications (Jan 2021). Driven by user feedback and technological advances they are constantly improving R2 and extending it with new options. Koster also conducts research in new ways of making complex data insightful. With their biology driven focus, they have been fortunate to be involved in many collaborations with research groups around the globe, which has already resulted in authorships on 30+ high impact publications.
After having been educated as a molecular biologist (Utrecht University), and subsequently trained as a cell biologist at the Netherlands Cancer Institute (NKI) (supervised by Prof. A. Sonnenberg), Jan Koster switched career to the at that time developing field of bioinformatics in 2003. He joined the team of Prof. Versteeg, and in 2007 obtained a staff position within the department with his own team of bioinformaticians. His group currently consists of 6 bioinformaticians. The focus of Koster’s group is on applied bioinformatics, where they try to bridge the gap between the molecular biologists and the complex data that they are generating using high throughput technologies. Aside from helping the biologists with the processing, analysis and interpretation of HT data, they have heavily invested in an idea conceived in 2005. ‘Wouldn’t it be great if molecular biologists could analyze and explore their own data’. With this aim in mind and their biology background and mindset they have developed the web-based R2 genomic analysis and visualization platform (http://r2.amc.nl). R2 is composed of a dedicated database containing the HT data, coupled to a web-based suite of analysis and visualization tools that can be operated by users without bioinformatics training. R2 handles many types of genomics data from mRNA gene expression to whole genome sequencing data. The success of this concept can be illustrated by a user base of 140,000+ users (IP-addresses) from around the world, ~2,000,000 page views a year and over 1,500 citations in PubMed listed publications (Jan 2021). Driven by user feedback and technological advances they are constantly improving R2 and extending it with new options. Koster also conducts research in new ways of making complex data insightful. With their biology driven focus, they have been fortunate to be involved in many collaborations with research groups around the globe, which has already resulted in authorships on 30+ high impact publications.
Kim-Anh Le Cao
 Dr Kim-Anh Lê Cao develops novel methods, software and tools to interpret big biological data and answer research questions efficiently. Kim-Anh is committed to statistical education to instill best analytical practice. She has taught numerous statistical workshops for biologists and leads collaborative projects in medicine, fundamental biology or microbiology disciplines.
Kim-Anh has a mathematical engineering background and graduated with a PhD in Statistics from the Université de Toulouse, France. She then moved to Australia first as a biostatistician consultant at QFAB Bioinformatics, then as a research group leader at the biomedical University of Queensland Diamantina Institute. She currently is Associate Professor in Statistical Genomics at the University of Melbourne. Kim-Anh has secured two consecutive NHMRC fellowships, published more than 90 articles and secured more than 8.5 million in competitive funding. In 2019, she received the Australian Academy of Science’s Moran Medal for her contributions to Applied Statistics, the Georgina Sweet Award for female scientists in quantitative biomedical science that recognize her contribution in multidisciplinary collaborations. She was selected to the international Homeward Bound leadership program for women in STEMM which culminated in a trip to Antarctica.
Dr Kim-Anh Lê Cao develops novel methods, software and tools to interpret big biological data and answer research questions efficiently. Kim-Anh is committed to statistical education to instill best analytical practice. She has taught numerous statistical workshops for biologists and leads collaborative projects in medicine, fundamental biology or microbiology disciplines.
Kim-Anh has a mathematical engineering background and graduated with a PhD in Statistics from the Université de Toulouse, France. She then moved to Australia first as a biostatistician consultant at QFAB Bioinformatics, then as a research group leader at the biomedical University of Queensland Diamantina Institute. She currently is Associate Professor in Statistical Genomics at the University of Melbourne. Kim-Anh has secured two consecutive NHMRC fellowships, published more than 90 articles and secured more than 8.5 million in competitive funding. In 2019, she received the Australian Academy of Science’s Moran Medal for her contributions to Applied Statistics, the Georgina Sweet Award for female scientists in quantitative biomedical science that recognize her contribution in multidisciplinary collaborations. She was selected to the international Homeward Bound leadership program for women in STEMM which culminated in a trip to Antarctica.
Patrick Deelen
 I am a post-doctoral researcher working in the Departments of Genetics of the University Medical Center Utrecht & the University Medical Center Groningen. I have Bachelor’s degree in bioinformatics from the Hanze University Groningen and a Master’s degree in bioinformatics (cum laude) from VU Amsterdam. I am primarily interested in improving the diagnostic yield of genome sequencing of patients who have a rare disease. I do this by integrating population genetics and transcriptomics.
I am a post-doctoral researcher working in the Departments of Genetics of the University Medical Center Utrecht & the University Medical Center Groningen. I have Bachelor’s degree in bioinformatics from the Hanze University Groningen and a Master’s degree in bioinformatics (cum laude) from VU Amsterdam. I am primarily interested in improving the diagnostic yield of genome sequencing of patients who have a rare disease. I do this by integrating population genetics and transcriptomics.
Gennady Roshchupkin
 Gennady is leading “Computational Population Biology” research group in departments of Epidemiology, Radiology and Nuclear Medicine at Erasmus MC Medical Center Rotterdam. He develops and applies methods for the integrative analysis of large-scale biological, epidemiological and clinical data. The group uses the latest approaches in genomics, medical imaging, computer science, statistics and machine learning to sort through increasingly rich and massive amount of data. The goals are to improve the understanding of how various omics affect the complex traits and to make use of such insights to improve the diagnosis, prevention and treatment of diseases whenever possible.
Gennady is leading “Computational Population Biology” research group in departments of Epidemiology, Radiology and Nuclear Medicine at Erasmus MC Medical Center Rotterdam. He develops and applies methods for the integrative analysis of large-scale biological, epidemiological and clinical data. The group uses the latest approaches in genomics, medical imaging, computer science, statistics and machine learning to sort through increasingly rich and massive amount of data. The goals are to improve the understanding of how various omics affect the complex traits and to make use of such insights to improve the diagnosis, prevention and treatment of diseases whenever possible.
Erik van den Akker
 I have a keen interest in studying the ageing hematopoietic and immune system and its effects on health. I performed most of my (post)doctoral training within (inter)national collaborative projects NCHA, IDEAL and BBMRI that aimed to elucidate the determinants contributing to healthy ageing. During this period, I specialized in developing methods for the integrative analysis of omics datasets within cross-disciplinary research. I'm currently working as an assistant professor on various computational topics, including the development of broadly applicable omics predictors for clinical outcomes and risk factors. In parallel, I perform research on the ageing hematopoietic system and its age-related changes to pre-leukemia, i.e. ‘age-related clonal hematopoiesis’, as highlighted by papers in Blood (Van den Akker et al. 2016) and Leukemia (van den Akker et al. 2020). Interestingly, age-related clonal hematopoiesis not only predisposes to hematologic malignancies, yet is also associated with a higher incidence of various low-grade inflammatory diseases, i.e. cardiovascular disease, and outcomes, i.e. cardiovascular mortality. I was awarded a Veni personal grant of the Dutch Research Council in 2019 to study the heterogeneity in clonal-hematopoiesis related outcomes using omics platforms. I was invited to the management team of the newly established Leiden Center for Computational Oncology (LCCO) to develop omics predictors for diagnostic and prognostic applications in acute myeloid leukemia.
I have a keen interest in studying the ageing hematopoietic and immune system and its effects on health. I performed most of my (post)doctoral training within (inter)national collaborative projects NCHA, IDEAL and BBMRI that aimed to elucidate the determinants contributing to healthy ageing. During this period, I specialized in developing methods for the integrative analysis of omics datasets within cross-disciplinary research. I'm currently working as an assistant professor on various computational topics, including the development of broadly applicable omics predictors for clinical outcomes and risk factors. In parallel, I perform research on the ageing hematopoietic system and its age-related changes to pre-leukemia, i.e. ‘age-related clonal hematopoiesis’, as highlighted by papers in Blood (Van den Akker et al. 2016) and Leukemia (van den Akker et al. 2020). Interestingly, age-related clonal hematopoiesis not only predisposes to hematologic malignancies, yet is also associated with a higher incidence of various low-grade inflammatory diseases, i.e. cardiovascular disease, and outcomes, i.e. cardiovascular mortality. I was awarded a Veni personal grant of the Dutch Research Council in 2019 to study the heterogeneity in clonal-hematopoiesis related outcomes using omics platforms. I was invited to the management team of the newly established Leiden Center for Computational Oncology (LCCO) to develop omics predictors for diagnostic and prognostic applications in acute myeloid leukemia.